Preamble — This post is a more detailed dive into the role of graphs in biomedicine and search in general which was outlined in a previous article on drug repurposing using AI.1 The topic of graph machine learning for relational data prediction has rich technical detail which cannot be covered sufficiently in a brief article. Graphs are a more generalized form of the attention mechanism of transformers that have been popularized by LLMs and the methods used to learn from graphs span a more diverse space of relational analysis which is particularly relevant for the heterogeneous data types in biomedicine.
More at: jasonsteiner.xyz
The Power of AI for Search
One of the most significant applications of AI is "Search". This is capitalized because it represents a spectrum of ideas. Training AI systems involves searching for a path toward minimizing a loss function, LLMs and other generative AI models involve creating a search distribution from which to generate new samples, and embeddings in vector databases are used to search for similarity to user queries in retrieval augmentation (RAG) systems.
There are two primary ways of thinking about search:
Search as a method for uncovering information existing within a system e.g., in the familiar Google search for content on the internet, or more recently, large-scale RAG engines like Perplexity.ai.
Search as a method for exploring new territory as guided by rules or boundaries e.g., Monte Carlo sampling, evolutionary recombination, or other methods that are employed in models like the game-playing Alpha series by Deepmind or Q*2
In each case, the utility of AI is to explore a very large space most efficiently and return useful information from that space -- whether a recommendation, an unearthed pattern or discovery, or a reinforcement pathway that provides the most desirable rewards.
I have written previously about the most powerful near-term applications of AI being in Knowledge Management3 with a particular emphasis on the biomedical field (although the topic is much broader).
In this field, there are three dominant forms of data: graphical, textual, and tabular/relational. There are powerful embedding and search methods for the first two -- namely convolutional neural network vector embeddings for images and LLM embeddings for text -- that can use relatively straightforward cosine similarity and vector databases for queries and responses.
The latter category of relational or tabular data is more complex, yet it is among the most dominant and powerful formats of data that exist and can encompass the two former categories in addition to a wide range of other data types.
This article will focus on applications of AI for relational search and how this will fundamentally change the way we interact with data through GQL — Graph Query Language4. I will focus primarily on how these tools can be used in biomedicine, however, the scope applies to relational data in general.
TLDR — SQL and tabular formats have dominated relational databases but queries and insight can be limited to the scope of existing data. Graph representations of relational data enable complex search and predictions across incomplete knowledge bases, significantly expanding potential insight.
Excel to Snowflake - Data Stored as Data
A significant portion of biomedical data is in Excel spreadsheets notwithstanding recent advances in bioinformatic infrastructure tools that are being developed by many startups. Data types range widely, metadata is abundant, often complex and lacking in standardization, and data is generated often piecemeal in rapidly iterated experiments in labs around the world.
A host of companies have cropped up with the primary goal of making biomedical and bioinformatic data more manageable. It is a requirement that companies have a data management system in place from day 1 and for the most part, this consists of developing standard protocols, data formats, database and data warehousing infrastructure, etc. When a company moves from Google Drive to AWS, GCP, Snowflake, (or other database providers), it's an important decision.
However, despite all of these data piping improvements, the underlying format of the data has remained largely unchanged. The predominant format remains tabular and the predominant search language for such data is SQL (Structured Query Language) which is used to pull, combine, transform, and interrogate the facts and figures in the underlying tables. This has been an extremely powerful tool to date and one that even the largest databases in the world rely on, including tools like BigQuery from Google — indeed, the original objective of the team that started Perplexity.ai was to develop a natural language to SQL tool to facilitate complex database queries5 for general users.
Excel to Pinecone - Data Stored as Representations
One of the most powerful capabilities of AI is the ability to represent data as vector representations - a numerical translation that enables calculations. The most common application of this currently is with retrieval augmented generation (RAG) using LLMs. This method takes (generally text-based) data, splits it into chunks of perhaps a few thousand characters and uses an LLM embedding model to convert those chunks into vector embeddings that are stored in databases provided by vendors like Pinecone. In this way, when there is an LLM query a RAG system can compare the embedding vector of the query to the embedding vectors in a database to determine which chunk of text is most closely related and then return that chunk as context to the query. This process is not too dissimilar from a more standard SQL query with an upfront transformation of the text embedding and a cosine similarity calculation.
Importantly, this type of embedding and relational calculations can be extended to any range of data types. In biomedicine, this is used most frequently to build heterogeneous knowledge graphs (KGs) that represent different types of data (for example, genes, drugs, diseases, clinical notes, ontologies, etc.) as vector representations and different types of relationships (for example, "binds to", "treats", "is a part of", etc.) as edges between these nodes.
Conceptually, the most important aspect of representation-based embeddings for information storage is the ability to perform continuous calculations of relationships, not solely discrete ones. With SQL queries, for example, one cannot go beyond a discrete analysis and return of the data that already exists. With embedded representations, it is possible to return continuous information, for example, as a similarity metric. In simple terms, we are moving from a paradigm of "does A co-occur with B" to a paradigm of "how closely does A co-occur with B", for any arbitrary A and B.
(Note: For clarity, this type of analysis is certainly possible with numerical data already -- the point here is that it is being extended to a wider range of data and relationship types).
From Tabs to Graphs
Information is often most useful when expressed in relationships, yet the prevailing methods of tabular data management have limited this. Take the example below — the question posed here is whether there is a dependency between Elements A and F. It can be rather difficult to distill with data represented in tabular format as on the left but becomes much more intuitive when viewed as a graph as on the right.
Figure: Ref from Neo4j6. Consider the challenge of determining whether Element A has a dependency on Element F in a tabular format or a graphical format
This concept seems relatively straightforward, but when considering the vast amount and diversity of data that exists in tables, the challenge of extracting relational information can be significant. For anyone who has used tools like AirTable and had the need to align the database on "key" value to combine "queries" ensuring that the keys flow through all relevant tables seamlessly, this conversion of the underlying data to a graph might provide a more flexible perspective.
Interpreting Graphs through Embeddings and Transformations
While graphs can be a visually appealing way of displaying information, their interpretation can pose significant challenges. To begin, they have no clear orientation or order. They are not spatially or sequentially oriented like images/tables or text. They may have long-range dependencies but they have no inherent symmetries. Further, with heterogeneous graphs, where different node and edge types may all have different properties, it can make tracing dependencies through graphs no simpler than through multiple database tables. This is where the tools of graph neural networks (GNNs) and embeddings become particularly useful.
The process follows two steps in general, which will be covered at a relatively high level here, that relate to the training of initial node and edge embeddings and a subsequent series of transformations for analysis and complex querying. For the interested reader, an excellent course on machine learning on graphs from Stanford is freely available online7.
A Brief Overview of Graph Embeddings
The central idea behind graph embeddings is to develop numerical representations of nodes (and edges) that capture the structural features of the graph. There are various methods to train these embeddings, but they can generally be distilled to the idea of sub-graph or local neighborhood similarities. In the case of node embeddings, this means that nodes that have a similar link structure to other nodes should be close in embedding space. Graphically this can be seen in the below example.
Figure Ref8. The most straightforward way of representing graph nodes as embeddings is based on their structural and neighborhood similarity. Nodes that are connected to common nodes embed closely. This image shows a network on the left and the color coded node embeddings on the right.
In a more general context, node embeddings can include a wide range of features about the nodes themselves (apart from just their link structure). For example, in biomedical data if one is embedding a "patient EHR record" using an LLM, the node embedding may represent patient metadata in addition to the LLM embeddings of the underlying data. The ability to capture complex information in a trainable embedding is one of the most powerful aspects of representation learning on graphs and can be used, in the case described for example, to uncover patient populations that might not otherwise be readily identifiable.
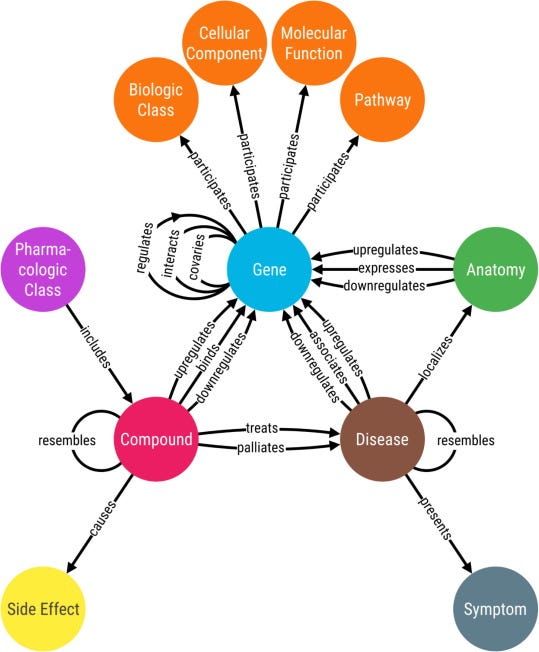
The simplest form of a graph embedding is one where nodes and edges are all of the same type (respectively) and the learned embeddings are representative of the structural feature of the network, however, in most data contexts multiple types of both nodes and relationships need to be considered. Such graphs are called heterogeneous graphs and they are the basis of most knowledge graphs. Such a graph may look like the below.
Figure Ref9. Biomedical knowledge graphs have multiple different types of nodes and edges that represent diverse information.
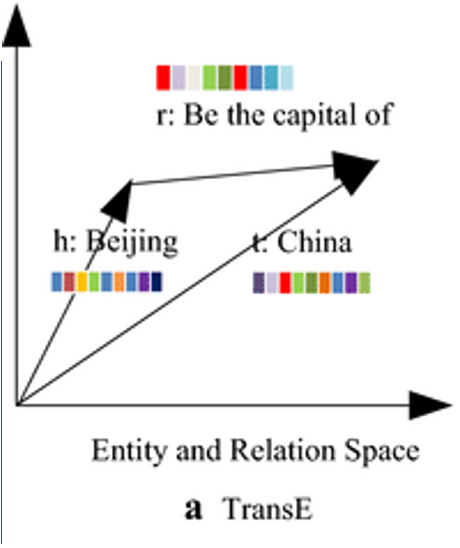
When constructing heterogeneous KGs like those in the figure above, it can be challenging to follow a reasoning path, for example, about how a particular entity is related to another entity through a given relationship type. When learning an embedding model for the nodes, there is no guarantee of an interpretable relational dimension between nodes. To enable this type of relational understanding, additional transformations can be performed that can integrate an embedding space of entities (or nodes) with an embedding space of relationships. One of the more popular embedding models is called TransE — this model creates embeddings of both nodes and edges in the same space such that their composition is accurate. A common example of this is for example, embeddings of capital cities and countries where the capitals and countries are embedded in relation to each other such the connecting vector represents “is capital of” relationship type.
Figure: Ref10 showing a vector composition between two entities (head and tail) and a relationship embedding (r) in the same space.
This type of embedding enables one to start with a given node and add any arbitrary number of “relational” edge vectors to determine what might be in the neighborhood of the composition. For example, any arbitrary city composed with “capital of” vector should result in the corresponding country.
A critical aspect of this approach is that the underlying data need not be complete in an underlying database. Being able to compose embedding queries as vectors in this space results in query compositions that can be compared to other embeddings via standard cosine similarity calculations which can result in continuous predictions. For example, if the city “London” is not directly linked to the country “England” in an underlying database, a composition of “London + capital of” embedding can be compared to an “England” embedding to determine the probability that it is a correct answer.
Importantly, these embeddings can be used to conduct multi-hop queries by simply composing multiple relationship embedding vectors to find nodes at the endpoints.
Query2Box Extends TransE to Complex Queries
An expansion of this concept for improved predictive results is known as Query2Box11. This is a method that builds on embedding models like TransE for more expressive predictions around compositional query neighborhoods. Starting from any node, one can then query any set of edge relationships by adding the edge embedding vector to the node and establishing a box "neighborhood" around the endpoint that captures any nodes within the box as likely to exist in relation to the starting node. Similarly, edge relationship vectors can be added to different starting nodes to span regions of their potential intersection. The process can be repeated with any number of relationship queries in succession to result in a final embedding neighborhood that spans the range of nodes that are likely to be answers to the query
An example of this from the Stanford lecture series on the topic is below:
Figure Ref12: Query2Box model for multi-hop or complex queries in embedding space.
In this case, one wants to find the answer to the following question "What drug causes shortness of breath and treats diseases associated with ESR2".
To accomplish this, one starts at an embedding for ESR2 and adds the relational edge embedding for “associated with”. This results in an embedding neighborhood for entities that are likely to satisfy this relationship. Taking a center point of this neighborhood and adding the relational embedding for “treated by” results in a new neighborhood of entities that are likely to satisfy the composed relationship.
Similarly, starting from the node for “shortness of breath” and adding relational embedding for “caused by” results in a specific embedding neighborhood. It is in the intersection of these neighborhoods where nodes exist that are likely to satisfy the full query.
Importantly these links do not need to be fully explicit in the underlying data as they would need to be with an SQL query. Operating in embedding space is able to reason about the likelihood of specific answers to complex queries which can provide significantly more insight.
GQL and the Future of Search
The most valuable near-term application of AI is in knowledge management. Significant businesses have built on the backbone of relational databases storing data in tabular formats and SQL remains a powerhouse of data science and analytics.
New developments in graph architectures, graph neural networks, and embedding models with GQL will fundamentally change the nature of what can be extracted from this assembled data. The most salient aspect is the ability to make predictions and reason over incomplete information. These techniques are being deployed across the vast data sets in biomedicine for applications like drug repurposing but the application scope extends to relational data in general.
We are entering a world of GQL. It'll be fascinating to see what's discovered in the vast troves of biomedical data.
For more on this and related topics: find me at jasonsteiner.xyz
References:
Drug Repurposing Using AI
Preamble: This post originally appeared on the Timmerman Report linked here and, being for a more general audience, is less technical than other posts on this Substack. However, it’s a very interesting and important trend nonetheless. One of the most powerful aspects of deep representation learning is the ability to express an arbitrary set of feature…
AI Mathematicians and Scientists
The Crazy Week at OpenAI The last week has been a tumult of news in the AI world - with the unexpected ouster of CEO Sam Altman from OpenAI, the pending implosion of its employee base, and the presumptive return to order with a revised board of directors and possibly corporate structure. It’s been a drama unfolding on X that has been unparalleled in rec…
Timmerman report: https://timmermanreport.com/2024/01/rebooting-ai-in-drug-discovery-on-the-slope-of-enlightenment/
Note: I am using GQL in this context in a more general term as a comparison to SQL. The terminology of Graph Query Language has recently (as of April 2024) been published as an ISO standard term for querying graphs. see https://www.iso.org/standard/76120.html
Ref: youtube.com/watch?v=LGuA5JOyUhE&t=1726s
Ref: youtube.com/watch?v=Ei-pYtYS6UY&t=1516s
Ref: youtube.com/watch?v=JAB_plj2rbA&list=PLoROMvodv4rPLKxIpqhjhPgdQy7imNkDn&index=1&t=8s
https://towardsdatascience.com/overview-of-deep-learning-on-graph-embeddings-4305c10ad4a4
https://www.sciencedirect.com/science/article/pii/S2001037020302804
https://www.researchgate.net/publication/340790855_Network_representation_learning_a_systematic_literature_review
https://arxiv.org/abs/2002.05969
Ref:youtube.com/watch?v=Nt66M2OsbCw&list=PLoROMvodv4rPLKxIpqhjhPgdQy7imNkDn&index=34