AI for Science
Primers on Building Agents
In science, if you know what you are doing, you should not be doing it
In engineering, if you do not know what you are doing, you should not be doing it
- Richard Hamming
TLDR:
In the theme of this Substack, this article is intended as an intuition primer. It is a semi-technical introduction for a general audience about topics related to deep learning and the sciences. This is a profound topic with a great deal of nuance and technical depth. If you find this type of article useful, please consider subscribing or reaching out to me at:
jason@jasonsteiner.xyz
jasonsteiner.xyz
This article is about science and machine learning, specifically deep learning. It will focus on the idea of ideas, the nature of information, and the process of learning. One of the central motivations for this article is to bring a better understanding of how AI will impact the sciences and our ability to interpret, model, and impact the world. I'll largely focus on the physical sciences with a particular emphasis on biology and the life sciences. I'll broadly consider "science" as a process of poking and prodding the world, constructing frameworks and theories about how it works, and conducting falsifiable experiments. This type of science exists on a continuum -- with one end being experimental and the other conceptual. These domains have different applications of deep learning which I will call "circuit search" and "policy search", respectively, that will be discussed later in the article.
Figure 1: Science is often divided into domains of experimental and theoretical. Deep learning has applications in both, but the most interesting is at the intersection. This article will discuss three main domains of deep learning in the sciences which I will call "circuit search", "policy search" and "agents". Circuit search is the application of deep learning to model data relationships, policy search is the application of deep learning to explore new conceptual territory, and agents blend the two to develop new science.
There is additionally a layer of consideration regarding "science" and "engineering". Historically these have been relatively separate domains as so eloquently stated by Richard Hamming (of Hamming encoding) in the introductory quote, however, the modern version of machine learning is blurring these lines. We should expect that increasing domains of science will be enabled by extensive engineering. Discovery is increasingly becoming a product of engineering infrastructure. For practical applications, there is a trend away from theoretical formulation of ideas and toward computational solutions. This is as much a practical necessity of the difficulty of solving complex systems, of which many are addressed via numerical methods, as it is the development of formal reasoning AI models that may, in fact, solve previously intractable symbolic expressions. Deep learning is approaching both sides of the scientific spectrum in meaningful ways -- and the most interesting is where they meet.
This article is somewhat lengthy so there are links to certain sections below.
Contents:
More Data, Less Progress
A few months ago, I downloaded my Google Takeout data. If you are unfamiliar, Google Takeout is a service that allows you to download all the user data that Google has collected on you.
It's eye-opening. It went back to ~2004 and contained the most unexpectedly detailed contents from the last 20+ years. The amount of data that has been generated over the past few decades is extraordinary. It's often thought that more data is a categorical good. In the life sciences, we have seen an explosion in data generation that has been led by technology platforms primarily in genomics (and increasingly other -omics), imaging, biosensors, the digitization of medical records, and the communication infrastructure to share scientific research. In physics, the field is characterized increasingly by the massive experimental scale of instruments like the Large Hadron Collider, which turns out petabytes of information per second. Two primary consequences of these trends are:
There's a tremendous amount of data
It's hard to tell if it's any good
When looking through the lens of academic publications (which form the majority of the public knowledge corpus), both of these are actually positive trends. The reward mechanism of publishing is novelty, not utility. To be sure, this is not intended to indicate that publications are not of value writ large, however, the pursuit of novelty for novelty's sake is not the objective of science. It is possible to produce infinite novelty with very little meaning. This is often masked by the statistical fact that the ability to find significance increases with variables and data quantity. This has resulted in a glut of information and publications that have unclear relevance and the propagation of the practice of p-hacking.
One consequence of this in the life sciences is manifested by the observation of "Eroom's" law -- or the idea that despite the dropping costs of data acquisition, the cost of developing drugs has increased. To be sure, there are many facets to this issue, but principal among them is that data has been generated where it is most accessible but often the least relevant. For example, generating data in cell lines that have low translational value.
A second consequence is that the sheer quantity of information nearly guarantees that some hypotheses will be validated. I spent some time working with several genomic foundation models with perturbation tasks to determine whether synthetic lethal genetic combinations could be prioritized. With the caveat that the models were not specifically fine-tuned, the resulting gene lists were assessed for their relevance in cancer biology. Essentially every gene had a hit in the literature. The quantity and quality of published data is such that almost any gene can be found to correlate with cancer biology in some publication or another. The resulting output did nothing to specify or narrow a search. This does not advance understanding but is the driver of a great deal of content in the scientific media. Very high sensitivity, very low specificity.
The two most important questions to begin any scientific inquiry with (including AI science) are the following:
What are the characteristics of the data underlying the question to be asked
What question to ask
The tools and approaches derive from these questions.
Framing Deep Learning
Before getting into more details, it's helpful to set a framework. Deep learning has been a resounding success but remains opaque to many people. Below is a brief framework with a few of the key ideas:
Deep learning can be thought of as a hierarchical extension of regression models, where layers of linear transformations are combined with non-linear activation functions to model complex, non-linear relationships in data. It is primarily the size of the layers and the number of parameters that have enabled the success of the approach in recent years.
"Training" is the process of optimizing the variables of the model to achieve a specific output given a specified input. This optimization is done by evaluating the derivatives of the output with respect to each variable and adjusting the variable along a gradient that moves the output toward the target. This can be thought of as a "search" for the optimal variable settings.
Deep learning can be thought of in 2 search modes.
Gradient Descent -- this is a mode that looks to minimize the difference between a model's output and a training sample - called a "loss function"
Gradient Ascent -- this a mode that is looking to maximize the expected value of a model's behavior or a reward function -- called a "value function"
These two modes can be mapped onto two major disciplines in deep learning which I will call:
Circuit Search
Policy Search
Each of these modes operates in a different computational framework that sets expectations on what may or may not be possible for training a network.
Circuit Search operates largely in the space of NP-complete - what this means is that it is not guaranteed that a solution can be found, but it is easy to check that a solution is valid. (technically, this is an optimization problem, not a decision problem so training is not in NP, but it can be reformulated to a decision by setting a loss threshold as opposed to an optimum to be in NP-complete)
Policy Search operates largely in the space of NP-hard -- what this means is that it is not guaranteed that a solution can be found nor is guaranteed that a solution can even be verified (both constraints in polynomial time)
In the context of AI for Science, we will discuss these two frameworks and their applications across the continuum of scientific inquiry (see Figure 1).
A Brief Description of Circuit Search
The view of gradient descent as the search for an optimal "circuit" can be a useful interpretation to link the theoretical and numerical aspects of science. It is a natural companion to scientific computing. Neural networks can be framed as circuits because of the use of activation functions like ReLU. These enable individual nodes to be either "on" or "off" depending on their input which is the basis of the Boolean logic.
Figure 2: A neural network can be thought of as an assembly of switches where each neuron is either on or off. More description about this is here: -- Programming with Data1 (https://biotechbio.substack.com/i/146329152/programming-with-data)
Training a deep neural network can be thought of as a process of finding a circuit that satisfies a set of numerical constraints. In the context of scientific computing, these constraints are typically set in a deterministic way as a set of differential equations that need to be satisfied in a physical system. Examples of this are the Navier-Stokes equations for fluids, flux balance equations for simulations of metabolic pathways, or genomic perturbation analysis on transcriptional cell states. Numerical solvers take a discrete incrementing approach to solve the next increment that satisfies the predefined constraints. This approach is computationally expensive, and can be near-term accurate, but is not purely deterministic because of the sensitivity of many systems of differential equations to initial conditions (e.g., leading to chaotic behavior). This sensitivity to initial conditions remains a barrier to long-term forecasting of complex systems in general, independent of whether they are calculated numerically or modeled with neural networks. Practically this means that our ability to make predictions about the long-term evolution of complex systems will be limited2. This is particularly challenging if systems have multiple stable points or transition states like the differentiation of stem cells, the progression of weather systems, or the evolution of cancer.
Training a deep neural network takes the opposite approach of numerical computing. Instead of predefining the symbolic equations that govern a system's evolution and calculating the process forward, the aim is to approximate what the aggregate calculation of such equations would be given an observed input and output. This aggregate calculation is expressed in the trained weights of a network. To express this differently, let's consider Navier-Stokes with a system of 5 partial differential equations. A neural network will express this solution in a string of simple terms. However, the aggregate of the solution to these 5 equations may require a neural network of millions or billions of multiplications, additions, and activations (or ReLUs) to accurately model the full system. The actual equation, if written out in long form, would be inscrutable, but the aggregate effect, if the coefficients are trained correctly will accurately map the inputs to the outputs that one might get from a step-wise numerical solver of the 5 main equations. This is amenable to many fields of science where the actual equations are not known -- notably the biological sciences. It is natural to think of biological systems as circuits such as the view of a metabolic network below:
Figure 33: metabolism is one of the most natural circuit interpretations in biology. There have been extensive efforts to model metabolism at different levels including whole-cell metabolism, but many of the variables remain unknown making the problem challenging to solve discretely.
When training a network, it's important that the inputs and outputs are correctly paired because the network will attempt to find the circuit that translates one to the other. In general, neural network training starts with a random initialization and then proceeds to gradually become refined. In many cases in science, we don't have to assume a random search space, because we know that there are some limits to physical phenomena which are represented by equations. These equations, for example, may be the activity rates of enzymes, or the physical relationships of force. One canonical example of this is the original AlphaFold protein models that, while based on neural networks, had a number of physical chemistry constraints, such as bond angles, that bound the scope of potential solutions.
For readers who are interested more in the circuit interpretation of neural networks and computability, the idea is closely related to the "satisfiability (sat) problem4" which asks for a given circuit of Boolean operations, is there an input that results in a True (or 1) output? The relationship of this problem to complex systems like biology is twofold: 1) Deep learning is a method of approximating a circuit that represents a given system and 2) Given that circuit, is there a way to produce a desired output — e.g., can we engineer biology? These problems are NP-complete so it is not guaranteed that any solution can actually be found, but we can verify if one is.
Figure 45: An example of circuit satisfiability -- in this case given the circuit shown, the input of 1's does not satisfy the circuit. The general question is whether there is any input sequence that produces a 1 at the output.
Thinking about deep neural networks as circuit solvers (or optimizers) provides a useful reference frame for bridging the availability of real-world data with the computational processing of neural networks. It frames the physical sciences as a computation problem, albeit one which may not be computable. For completeness, there are important caveats to this analogy including the fact that neural networks need not be binary switches, and inputs/outputs may be continuous, but the computation framework is the same.
A Brief Description of Policy Search
While deep learning for circuit search can be thought of as a relatively bounded problem -- deep learning for policy search does not necessarily have this property. Policy search deals with the question of what to do. These questions approach the scientific process from the right (see Figure 1). A general framing for these questions is: given a particular state, a particular set of possible actions, and a set of probabilities about what those actions may result in, what is the optimal function that maps states to actions to obtain a desired outcome (called the reward)? This formulation can be straightforward in the context of games that are based on rules and have definitive outcomes, such as chess and go, or highly ambiguous such as optimizing the outcomes of the scientific method.
Policy search models are generally characterized by several strong assumptions such as the Markov assumption, a framework where the next step only depends on the current state and none of the history of past states. Decision frameworks that include probabilities of any action leading to a subsequent state and the value of those actions collectively make up a "policy" function. The objective of a policy search is to determine the best actions to take based on a given state that will optimize the cumulative future value. Visually this looks like a branching tree starting from a point where each path is characterized by the function that maps states to actions to reward. A nice visual from Toward Data Science of what this type of tree looks like for tic-tac-toe is below — there are some instances of recursion for example where board states can be visited multiple times via different paths.
Figure 56: An example of a decision tree for tic-tac-toe which assigns values to states and can be used to calculate optimal policies for actions in any given state
Any policy that is tested -- e.g., do Y in state X is used at every step of the decision tree which allows for cumulative probability of reward from any initial state. A basic training setup is to have training samples which are starting states (and the actions possible from each state), a randomly initialized neural network that represents the mapping of a state to a reward (or some version of this), also known as the policy, and then calculating forward any number steps the expected value of the given policy. Training updates the weights of the neural network to optimize the expected value of this reward.
An example of how this might be used in language-based reasoning is the following: Reasoning in natural language can be thought of as a progression of connected statements. Each statement is a state and the action at each state can be thought of as a continuous set of actions that result in the next state -- which is the next statement in the reasoning process. For example, if A then B, if B then C --> if A then C. In the domain of language, each of these states can be represented by its embedding -- this is simply a point that exists in high dimensional space that represents the sentence. The action space for the next statement is the possible set of vectors that translates to the embedding of the next statement -- and the trace between these states, and all subsequent states forms one reasoning thread. Deep neural networks can be used for many of the mapping functions -- for example, to calculate the translation of an initial embedding point to the next point along the trace. The weights of a network can be continuously tuned to model this mapping toward an optimal reward state -- this becomes the policy of the network.
Let's say for a reasoning task a correct answer is "C" and we start from a state "A" and move to a new state "if A then B”. We train a model to develop a policy that will eventually get us from A to C. So, we start at "if A then B" with a random set of weights in our neural network -- the space that can be mapped from A is, let's say the continuous embedding around "if A then B". This is not necessarily true for models like o-1 because the auto-regressive completions of the underlying LLM would constrain the possible embedding states of the subsequent statements and we can approximate that policies of nearby embeddings would be similar, but let's say for this example it is naive. An agent could explore this space around A using a policy (which would be a neural network that converts embedding vector A to another embedding. It could then iterate this policy over time steps and the agent would end up at some end point, for example, X. X is far from C so the reward for that policy is low. Updating weights to get closer and closer to C, ultimately the model may converge on a policy that maps the statement "if A then B" to the embedding for B and the same policy applied iteratively which produces "if B then C". So, we can arrive at a point "C" which would have a high reward. In this simple version, the policy might have the interpretation of "translate to the next letter of the alphabet". Applying this policy iteratively leads to higher reward paths. It's possible to find a policy that might look like "move forward three letters and backward two". This policy could also optimize the trajectory. In practice, state spaces may not be continuous so only certain policies are permitted. This might be the case, for example, in mathematical theorem proving where the formulation of a theorem must meet certain criteria. In natural language, the reasoning space might be very continuous and very large.
Figure 6: Reasoning policies can be thought of as exploring a search space around a starting embedding. In general, for this type of reasoning policy search, there must be a way to quantify the expected rewards or values of each state. For formal systems, this can be explicitly defined as correct or not, but for more ambiguous tasks like reasoning a scientific theory, there may be degrees of correctness that are necessary to assign in order to train a policy. In this diagram, a high reward policy might be “statements that advance one letter in the alphabet which gets you from A to C through B. Rewards for policies that end up at X are low, policies that move closer to C are high.
The application of this type of policy search can also be rather diverse with respect to actions taken. They do not necessarily have to reside in a single embedding space - for example, a policy could be "if a given state has a confidence value below X, seek more information". In the context of scientific inquiry, this would be a natural process that a scientist might follow which then could progress to a new state with improved confidence scores where the policy is applied to that new state.
Increases in computing power have significantly enhanced this type of model by enabling very high dimensional state representations, continuous action spaces, and deep networks. This framework is very useful in systems that can be formalized (meaning they can be expressed in symbolic form) such as mathematical theorem proving, but also extends to natural language reasoning7.
The connection to science is direct and clear in the way theories, hypotheses, and experiments are designed. Scientific discovery is the process of reasoning about knowledge and creating the next step in the chain. One of the most fascinating questions about scientific thought is how the mind synthesizes and interprets a vast and diverse set of experiences and facts. The individual scientist is unique in this respect -- with their "mental policies" guiding one thought to another being shaped by their specific set of experiences and training. These are shaped by collaborations and social networks. Some proxies for thinking about this look at citation networks in publications to map how concepts have evolved or semantic analysis of publications to see where white space or connections may be emerging between disparate disciplines. There is also no doubt that serendipity plays a significant role, particularly for major breakthroughs like the discovery of antibiotics.
Progress in science is often very incremental -- a PhD is considered a small protrusion on the border of knowledge. If one were to be able to develop an effective policy model for scientific reasoning that maps decision processes, one could imagine these protrusions in the unknown could be rapidly accelerated with high success rates.
Figure 78: A protrusion on the frontier or knowledge. Generating a policy that could navigate the current reasoning landscape of scientific inquiry could accelerate the production and success of pushing these boundaries.
Science is as much a social enterprise as it is a technical one. Sharing ideas can be thought of as updating the policy networks of individuals by updating the value assignment of a given state (in the previous language example this would mean making a particular statement a high reward progression from its previous state). However, the opposite is also true -- misinformation or incorrect information leads to an expanding search space that diminishes the ability to effectively assess good policies. Novelty is not the primary objective of science.
Data for Circuits and Policies
The two frameworks above are conceptual, but both require data to develop and train. The type of data is often distinct — with one form being experimental and one form frequently being synthetic. In either case, the data should have the following characteristics:
It should be representative of the task at hand — this is a version of “garbage in, garbage out”
It should be transformed into a standardized format for comparison and calculation — avoid apples and oranges.
Deep learning has a powerful tool kit to achieve these ends when dealing with real world data that focus on translation and representation embeddings.
An embedding is simply a common representation of concepts, objects, etc. that allows for their comparison. In deep learning, they are vectors of numbers. However, the idea of an embedding is more general -- a commonplace example is money which can be thought of as an "embedding" of economic value. In this case, any given economic value is distinct -- e.g., making a podcast, selling a product, providing services, etc. -- but they can all be compared in a common framework of money. This is also a "compression" of the economic value provided -- meaning that it is not necessary to assess the entire value chain of a product -- it suffices to know the final price.
There are two steps to creating an embedding
translate information into a common format -- e.g., numbers
determine what those numbers should be
Examples of the translation aspects are relatively basic in practice. For language, it's done through "tokens" which are just collections of 3-5 characters that appear together in text. You could also just say that each word is a single digit and then say your translation of a word is just a 1 where that word exists and 0 everywhere else. This is called "one-hot" encoding and is very common. For example, DNA is one-hot encoded where every base is one of [1,0,0,0], [0,1,0,0], [0,0,1,0], [0,0,0,1] where the columns represent the bases A,T,G,C. An image might be one-hot encoded for each pixel if it is red, green, or blue as [1,0,0], [0,1,0], [0,0,1]. This type of translation doesn't add any value to the description of the information, but it does provide a way to express different types of information in a common format.
Now that everything is framed in the context of numbers, the second step is to determine how to represent the specific collection of numbers in a given order. For example, the sequence AATG is different than TGAA -- so these should have different representations. Naively we could just concatenate their translations as above -- those would be different and unique, but this doesn't give us much. The process of creating an embedding is typically one of compressing information to a smaller footprint which can carry all of the information of the original data but in a simpler manner. For example, in the English language, the letter "q" is always followed by the letter "u" -- as such it is not necessary to convey the "u" if you want to uniquely identify the "qu" character. The English language overall is about 50% redundant.
The process is typically done through an autoencoder. The idea is simple -- a series of calculations act on raw data with the objective of reducing the size of the data to a minimum size and then reconstructing the data from the compressed state. The calculations have variables that are adjusted based on how far the reconstruction is from truth. If you can reach a point where the reconstructed version matches the truth, then you know that all the information in the original data can be represented in a compressed state. This compressed state is the embedding. Importantly, because the size of the compressed state is arbitrary, you can translate a wide range of input sizes into the same compressed state. In language, for example, this means that sentences of different lengths can be compared equally. It also means that it is possible to compare, for example, the embedding of a picture, with the embedding of a sentence. The process of trying to get these two embeddings to match is the way that text-to-image models are trained -- when the two embeddings are the same, you can decompress the same values into either a sentence or an image that match each other.
Figure 89: Image of an autoencoder. If you can reconstruct the original data, you know that all its features are contained in the lower dimensional space -- this is the embedding.
The learning of embeddings is one of the most central aspects of deep learning. It's important to note, however, that embeddings are not fixed, they are a property of the model and the specific data. They are not generally translatable between models, and they have no specific relevance apart from their relative comparison within the same model architecture. One caveat — there are models that aim to learn mappings across existing embedding models without retraining to make them comparable10. Also importantly, embeddings do not have any specific meaning - -they are simply the product of a calculation. The meaning of embeddings is provided by the meaning of the input data. It is very possible to learn embeddings of meaningless data. Such embeddings can be used, manipulated, compared, and so forth, but do not have any meaningful representation in the real world. The importance of this point cannot be overstated in the context of AI applications for science. One of the primary avenues for the field is to develop higher-order characterizations via embeddings for increasingly complex systems. The most common approach is to simply concatenate the digital translations of different data types into a single input. For example, if the vector [1,0,0] represents Feature A and [0,0,0,1] represents Feature B of a shared entity, then to create an embedding of that entity, the approach would be to just use a concatenated vector of [1,0,0,0,0,0,1] as the input. This vector presumably contains the details of Feature A and Feature B and the compressed version of this would then represent 1) the compression of Feature A, 2) the compression of Feature B, and 3) the compression of the relationship between Feature A and Feature B.
Critically, when determining whether this embedding is valid, it's necessary to know that Feature A and Feature B are coming from the same source. It is not necessary to assume that Feature A and Feature B are directly related, however, the larger the intervening hidden variables connecting them, the more data will be required to map a meaningful relationship.
The idea of representing information in this embedding format allows for easy comparison between complex objects by simply comparing how close their embeddings are. Augmenting this idea of embeddings is the ability to direct models to learn embeddings that fall into certain distributions, for example, sparse embeddings or variational embeddings. These types of embeddings are used to try to understand the underlying features that are being identified in the compression -- this is central to the field of interpretability of neural networks where a particular dimension of an embedding may represent, for example, a specific noun or a specific sentiment like "happy", or "deceptive".
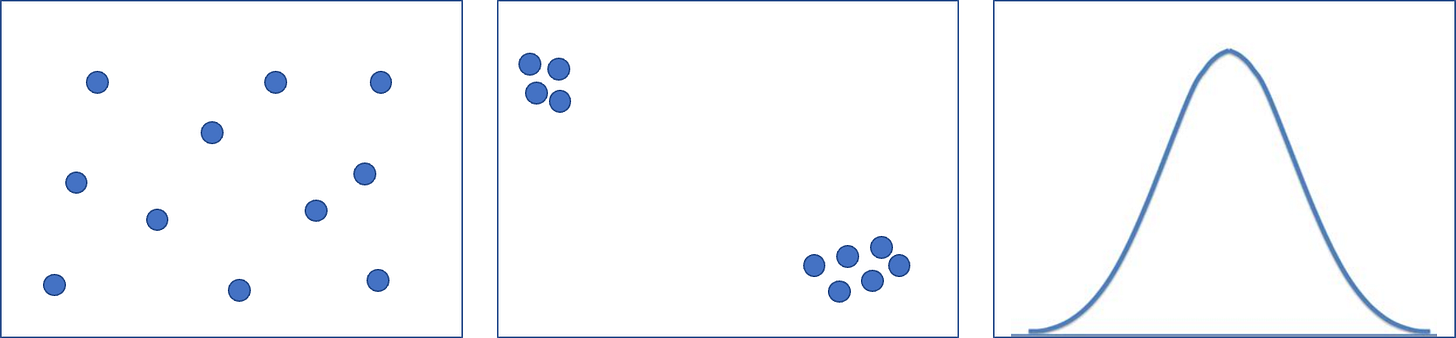
Figure 9: Examples of different embedding spaces -- on the left is a naive embedding space where embeddings can be anywhere that calculations permit, the middle is a sparse embedding where most dimensions are near zero so as to increase embedding separation and enhance interpretability, the right image is a variational distribution which forces embeddings into a distribution, typically Gaussian-- this format is common for generative model because you can sample new embeddings from the distribution to make valid images (if you sample from the embedding space of the prior two you will likely sample a nonsensical embedding.
This is a core pillar of deep learning -- and enables the encoding of information, the ability to compare different types of information, and the ability to predict or forecast expected relationships given existing ones-- the latter being the basis of autoregressive models for language generation or any number of "fill-in-the-blank" prediction models.
Examples of important embedding models are:
word2vec11 -- the original embedding model for words that was based on how frequently words co-appear
node2vec12 -- the original embedding model for nodes in a graph that represented how similar the nodes are to each other based on the graph structure
Universal Cell Embeddings13 -- one of the original embedding models for cells that aimed to aggregate several cell features into a common identity
Enformer14 DNA embeddings
ESM15 protein embeddings
Synthetic Data
Synthetic data deserves a mention but is more extensively covered in another prior article (ref 21). In brief, synthetic data can be very useful when it can be generated based on predefined models or rules. This covers many applications in the sciences that are characterized by equations or generative models that do not require specific grounding in fact (for example languages and images). Where these two criteria are not met, synthetic data is of much less value. There may be some cases where synthetic data as a negative control might have utility such as training GANs to distinguish real vs generated data.
In the life sciences, synthetic data is a much less viable option than in many other domains for “circuit search” problems for two main reasons:
Unlike the physical sciences, the equations that describe biological processes are not often well defined to create simulations.
Training data needs to be grounded in fact which makes generative approaches often less applicable. For example, it’s possible to use generative models to create new sentences or translational variations on images because they do not require specific factuality or can be generated by mathematical rules. In contrast, generating physically valid states of complex systems like cells or living systems is harder.
Synthetic data, however, does play a significant role in “policy search” for the practical reason that one cannot readily generate the scope of data necessary to explore all hypotheticals. This is more acceptable because policy searches in the sciences largely relate to conceptual topics like design, hypothesis generation, etc., which do not have the same constraints.
Generating Data
Embeddings are the center of deep learning as a generalized way of representing information. But just like any information, it remains essential to understand what is actually being represented by an embedding. Drawing the connection back to the idea of neural networks as "circuit search" -- the inputs matter for the wiring. It's straightforward to imagine that the size of the circuit is related to the complexity of the input/output relationships and consequently the amount of data that will be needed to effectively train a model16. This is particularly relevant for drug discovery where the goal is to find interventions that affect networks in desired ways (this is a version of the satisfiability problem in computation). One of the most important decisions is defining the level of data that you want to operate on and the tools for data acquisition that are available.
A specific example of this is generating data for the impact of small molecules on cells. One decision point could be to gather data solely at the phenotypic level via imaging. This type of data will not provide specific resolution for molecular mechanisms of action, but it may provide information on aggregate response. A data acquisition platform then pairs high content images with high throughput compound screening and is trained to map image embedding transitions to molecular embeddings.
Figure 1017: An example of higher-order embeddings. Pairing high-content imaging of different phenotypes in embedding space and aligning with molecular embeddings can produce a mapping of molecular structures to phenotypic changes. Chemical structures are commonly embedded using graph networks by constructing a "virtual node" that is connected to every atom in the structure and finding the embedding of that node. This model of pairing embeddings is a common one for data generation to train models.
The above is an approach for the types of data that might be useful to generate. Another aspect is the design of the experimental data sets for the most efficient learning process including the incorporation of prior knowledge into the design. For example, if we were to design data generation experiments for gene regulatory networks, it would be prudent to consider existing knowledge on gene expression to design experiments that are optimally information-rich18.
The key to data generation is having clarity on the size of the “circuit” that you are aiming to map. If the mapping is relatively straightforward — e.g. mapping chromatin accessibility to gene expression, the required data to map the circuit is scaled to match. If the mapping is very large, for example forecasting the effects of a compound in the body during clinical trials, the data requirements may be very large. In practice, most ML models in the sciences fall somewhere in the middle and aim to address pieces of the workflow like whether a compound is likely to be toxic or to bind a particular protein.
Data for Policy Training
Much of the content around data in the sciences is focused on experimental generation, however, there is also a consideration for training data generation of decision policies for agents. It is of note that many agentic models are trained on synthetic or augmented data, including the Alpha series and MuZero19 from DeepMind and the Cosmos20 engine from NVIDIA21.
A more detailed discussion of data for policy training in the life sciences is content for a future article.
Closing Notes
The computational paradigm of generation + search + reinforcement combined with increases in processing capacity will have a significant impact on the sciences. Regardless of the computational limits of complexity, one need not find an optimization to make progress. The central issues surrounding how this will evolve are the same as in any scientific inquiry -- specifically to be clear about the scope and design. The difference is that scope and design in the context of deep learning and AI are distinct from the practices of most scientific disciplines. It will be the convergences of these paradigms that will create the next stage of scientific super intelligence.
Additional Learning
In addition to the references cited directly in this article, I would recommend the following for additional education on these topics.
Steve Brunton has an excellent YouTube series on physics informed neural networks — https://www.youtube.com/@Eigensteve/playlists
Mutual Information has one of the clearest and cleanest series on reinforcement learning and agents I have found — https://www.youtube.com/@Mutual_Information/videos
References
https://biotechbio.substack.com/i/146329152/limits-on-computability-of-sequences
https://en.wikipedia.org/wiki/Boolean_satisfiability_problem
https://www.assemblyai.com/blog/content/images/2022/01/autoencoder_architecture.png
https://arxiv.org/html/2310.03320v4
https://arxiv.org/pdf/1301.3781
https://arxiv.org/pdf/1607.00653
https://www.biorxiv.org/content/10.1101/2023.11.28.568918v1
https://www.nature.com/articles/s41592-021-01252-x
https://www.science.org/doi/10.1126/science.ade2574
Data Entropy and AI Reasoning
TLDR: This article discusses data and topics around AI reasoning including pairing generation with search and reinforcement and the recent release of o1 from OpenAI.
https://arxiv.org/pdf/1911.08265
https://www.nvidia.com/en-us/ai/cosmos/
To Be Or Not To Be Synthetic: Data In Bio x ML
Summary: Synthetic data is becoming more central to the training and development of deep neural networks. This article describes some of the intuition around its utility in different domains.